Research Highlights from the Resource
Aplysia as an Alzheimer’s Disease (AD) Model
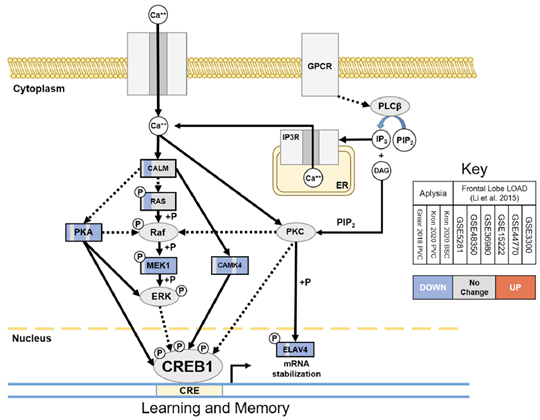
Molecular studies in the Resource of the effects of aging on the transcriptomes of sensory neurons revealed aging signatures that included metabolic, proteostatic, and neuro-synaptic impairments similar to those that also occur in AD and related dementias. Comparison of differential expression in three aging Aplysia sensory neuron data sets from the Resource with a meta-analysis-derived gene sets of human frontal cortex late onset AD (FL LOAD: Alzgset; Hue et al. 2017 and AlzGene; Bertram et al. 2007) identified 68 putative gene orthologs that were concordantly differentially expressed. Commonly upregulated genes included cellular stress-induced genes such as ANKZF1, BTG1, DDIT4L, and SSR1, as well as elements of the proinflammatory toll/interleukin receptor signaling pathways such as MYD88, NFKBIA, MAP3K8, and BIRC3. Commonly downregulated genes were representative of diverse processes including synaptic vesicle dynamics (SYN2, EXOC8, NAPG, SVOP, ARF3), transport of cellular cargo (DCTN6, KIFAP3, RAB6A), energy metabolism (GOT1 and 2, MDH1, CYCS, NDUFV1, PCCB), cyclic-AMP response element-binding protein (CREB)-mediated learning and memory (MAP2K1, PRKACA, CAMK4, ELAV4), and mitochondrial homeostasis (GDAP1,TUSC2). Figure 1 below is an illustration of the orthologs in a learning and memory pathway downregulated in common between Aplysia sensory neuron aging and FL LOAD.
|
Figure 1: Orthologs in learning and memory pathway downregulated in common between Aplysia sensory neuron aging and FL LOAD. Commonly downregulated genes included major kinases of CREB1 (PKA, CAMK4, MEK1) and ELAV4, which stabilizes mRNAs of CREB1 target genes. This suggests that CREB1 signaling disruption is a common cause of cognitive impairement in the 2 model systems.
|
Co-expression analysis identifies neuroinflammation as a driver of sensory neuron aging in Aplysia
Aging of the nervous system includes increased inflammation that results in cognitive impairment.
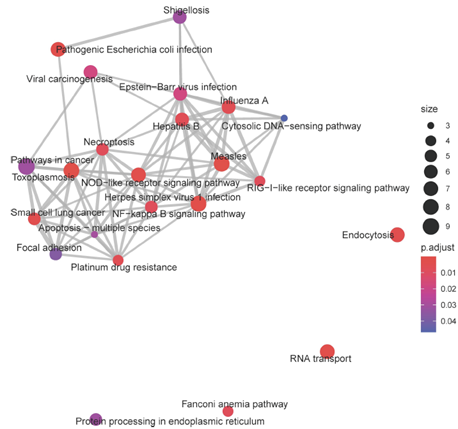
Coexpression analysis allows for identification of networks of genes and their putative central regulators. We used weighted gene correlation network analysis (WGCNA) to identify co-expression networks in aging Aplysia sensory neurons. Kyoto Encyclopedia of Genes (KEGG) analysis and investigation of central module transcripts identified signatures of metabolic impairment, increased reactive oxygen species, compromised proteostasis, disrupted signaling, and increased inflammation (see Figure 2).
Although modules with immune character were identified, there was no correlation between genes in Aplysia that increased in expression with aging and the orthologous genes in oyster displaying long-term increases in expression after a virus-like challenge (Lafont et al 2020). This suggests anti-viral response is not a driver of Aplysia sensory neuron aging.
|
Figure 2: Each node represents a KEGG pathway, with node size representing the number of transcripts annotated to that pathway, and color denoting the significance of that enrichment (brighter red is most significant). KEGG pathways with overlapping transcripts sets are connected by grey lines, or edges. Edge width is determined by the number of overlapping transcripts. Many of the significant pathways, with overlapping gene sets, are associated with immune activation, such as NOD-like receptor signaling, RIG-I-like receptor signaling, and NF-Kappa-B signaling pathway. The increasing expression trend of this module’s eigengene throughout aging suggests consistently increased activation of these pathways in aging.
|
Bertram, et al 2007. Nat Genet. 10.1038/ng1934
Kron and Fieber, 2021. J. Molec. Neurosci. 10.1007/s12031-021-01918-3
Kron and Fieber 2022. PLoS ONE https://doi.org/10.1371/journal.pone.0252647
Hu, et al. 2017. Alzheimers Res Ther. 10.1186/s13195-017-0252-z
Lafont et al. 2020. mBio. https://doi.org/10.1128/mBio.02777-19





